Wheat breeders have been using a lot of wild relatives to enrich the gene pool of wheat, especially on stress tolerance and disease resistance. Therefore, some wheat germplasms are alien Introgression Lines. QTLs identified in these regions in a biparental population will have a big confidence region, because there has no recombination. Two ways to overcome this:
Cross the resistant line with CSph1b to get homeologous recombination.
Create EMS mutants from the resistant parent and cross some of the loss-of-resistance mutants with the parent to create a new population. There will be recombination, and the EMS SNPs in that region can help screen recombinants.
The 2nd way might be easier and will introduce no other resistant genes. Now the new question is how to get the EMS SNPs for recombinant screening.
1.Do exome capture for both the wild type and the mutants. It should be enough for marker development, but may not capture the candidate gene, because resistance genes are usually new genes and may not be in the captures.
- RNA-seq to get all expressed genes in both the wild type and the mutants. Sequence several samples from different growth stages to make sure your candidate genes get included.
If we use the 2nd method, we might sequence one pool of the resistant sister lines and one pool of the susceptible sister lines, which might help fast pinpoint the candidate gene.
I think it might be better to do RNA-seq of the wild relative directly (if you know the line used for the introgression), which might be easier for De novo assembly, because you will not have the homologous genes.
Keep in mind: this is just my thought and I have not personally used this before.
The mutation method is similar to MutRenseq (Steuernagel et al. 2016, Nat Biotechnol 34, 652–655), but does not use the NLR capture, because not all R genes are NLR genes.
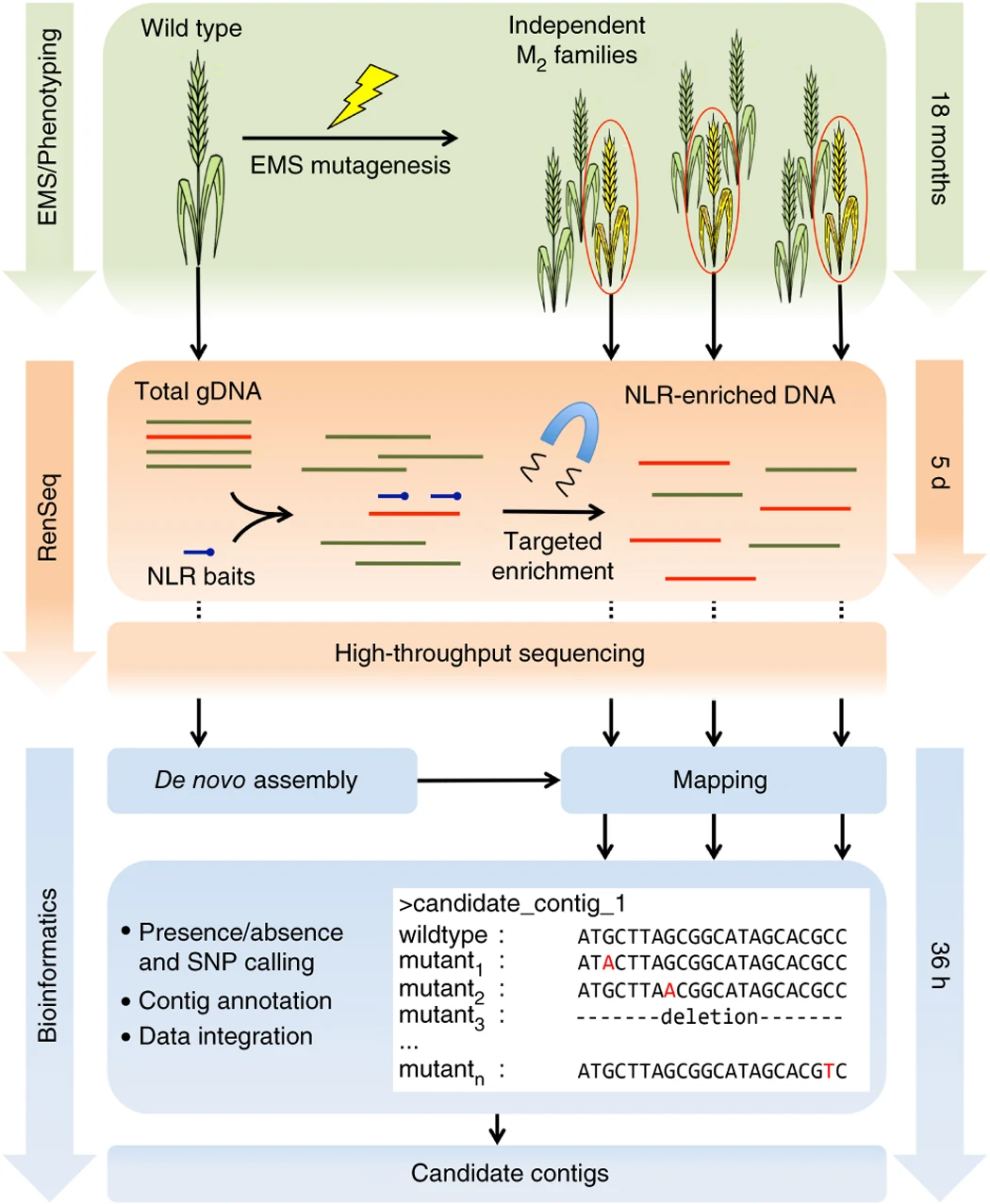
(Source: https://www.nature.com/articles/nbt.3543)