Lesson: Develop markers in the peak region FIRST!
I have been doing fine mapping of spikelet number QTLs in wheat for a while. I had found heterogeneous inbred families (HIFs), but I did not move very fast because I did not develop enough markers to genotype (or characterize) the regions of heterozgyous lines for the peak markers. So PLEASE develop at least 5 markers in the flanking regions from the beginning of the project! Then genotype the heterzogyous lines, find recombinants, and then do progeny test in the next generation, and go on!
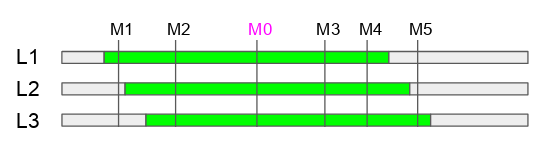
In the figure above, we have 3 HIFs that are still heterozygous for the peak markers M0. Green color means heterozygous and the gray color means fixed regions. To find HIFs, we planted about 10 to 20 seeds and genotyped them with the peak marker M0 first, and suppose we found 3 lines that are still segregating for the peak marker (Fig 1).
The heterozygous regions of these 3 lines are possibly different.
We need to develop a few markers between the flanking markers to find their heterozygous length. Please do this early, because it is easier to deveop markers than phenotyping. For example in the figure above, we know L1 is fixed for M5, L2 is fixed for M1 and M5, and L3 is fixed for M1.
Do progeny test for all the 3 lines, and we can find the gene location relative M1 and M5. If L1 is still segregating, we know the gene is not on the right side of M5; if L2 progenies are segregating, we know the gene is between M1 and M5.
In the progeny test, we can find more recombinants and keep going to narrow down the region. But the more markers, the better to define the heterozygous region.